Chimpanzees#

© Fauna & Flora
Recordings from:
Plooij, F. X. et al. (2015). An archive of longitudinal recordings of the vocalizations of adult Gombe chimpanzees. Scientific Data. https://www.doi.org/10.1038/sdata.2015.27
Setup#
[1]:
# import packages
import chatter
from pathlib import Path
[2]:
# set config parameters that depart from defaults
config = {
# spectrogram parameters
"n_mels": 128,
"fmin": 0,
"fmax": 4000,
# preprocessing parameters
"use_biodenoising": True,
"use_noisereduce": True,
"high_pass": 100,
"low_pass": 4000,
"threshold": 2,
"target_dbfs": -20,
"compressor_amount": -20,
"limiter_amount": -10,
# simple segmentation parameters
"simple_noise_floor": -60,
"simple_silence_threshold_db": -30,
"simple_min_silence_length": 0.00001,
"simple_max_unit_length": 2,
"simple_min_unit_length": 0.1,
# other parameters
"plot_clip_duration": 10,
}
config = chatter.make_config(config)
# initialize the analyzer with the configuration
# using only 8 cores to avoid memory crash during segmentation
analyzer = chatter.Analyzer(config, n_jobs=8)
model = chatter.Trainer(config)
# set paths
input_dir = Path("/Volumes/Expansion/data/chatter/examples/chimpanzee/recordings/raw")
processed_dir = Path(
"/Volumes/Expansion/data/chatter/examples/chimpanzee/recordings/processed"
)
h5_path = Path("/Volumes/Expansion/data/chatter/examples/chimpanzee/spectrograms.h5")
csv_path = Path("/Volumes/Expansion/data/chatter/examples/chimpanzee/spectrograms.csv")
model_dir = Path("/Volumes/Expansion/data/chatter/examples/chimpanzee/model")
output_csv_path = Path("/Volumes/Expansion/data/chatter/examples/chimpanzee/output.csv")
Using 8 cores for parallel processing
Initializing convolutional variational autoencoder
Using device: mps
Preprocessing#
[7]:
# demo the preprocessing pipeline
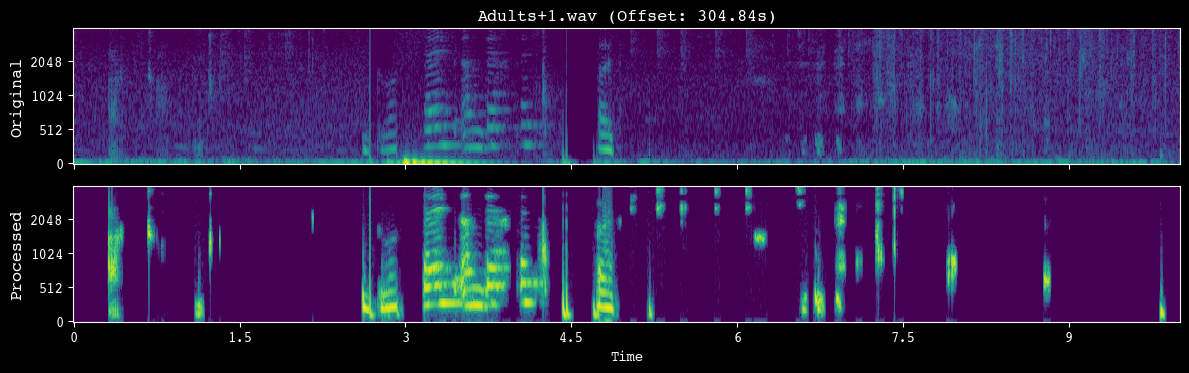
analyzer.demo_preprocessing(input_dir)
--- Demoing preprocessing for: Adults+1.wav ---
Segment: 304.84s - 314.84s
[8]:
# preprocess recordings
analyzer.preprocess_directory(input_dir=input_dir, processed_dir=processed_dir)
--- Found 7 audio files to preprocess ---
Preprocessing audio: 100%|██████████| 7/7 [03:08<00:00, 26.89s/it]
--- Preprocessing complete. Standardized WAV audio saved to /Volumes/Expansion/data/chatter/examples/chimpanzee/recordings/processed ---
Segmentation#
[9]:
# preview the segmentation pipeline
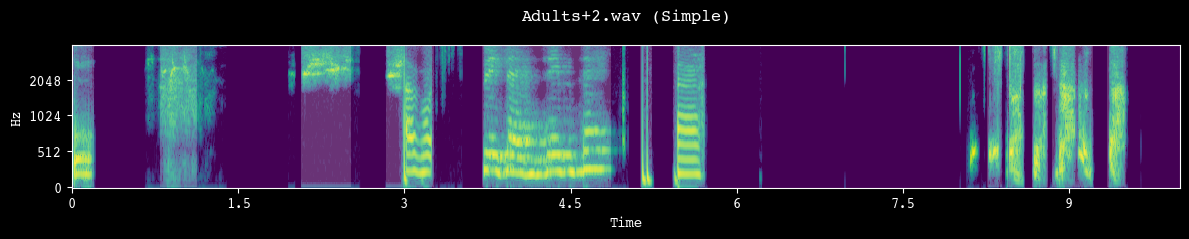
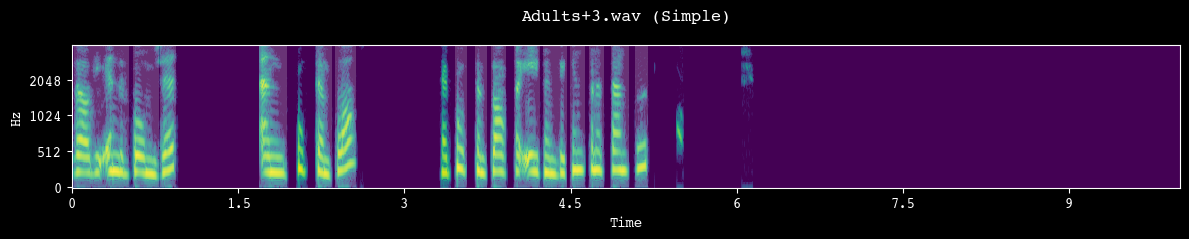
analyzer.demo_segmentation(input_dir=processed_dir, simple=True)
analyzer.demo_segmentation(input_dir=processed_dir, simple=True)
analyzer.demo_segmentation(input_dir=processed_dir, simple=True)
analyzer.demo_segmentation(input_dir=processed_dir, simple=True)
--- Demoing segmentation for: Adults+2.wav ---
Segment: 478.61s - 488.61s
--- Demoing segmentation for: Adults+3.wav ---
Segment: 439.83s - 449.83s
--- Demoing segmentation for: Adults+1.wav ---
Segment: 188.92s - 198.92s
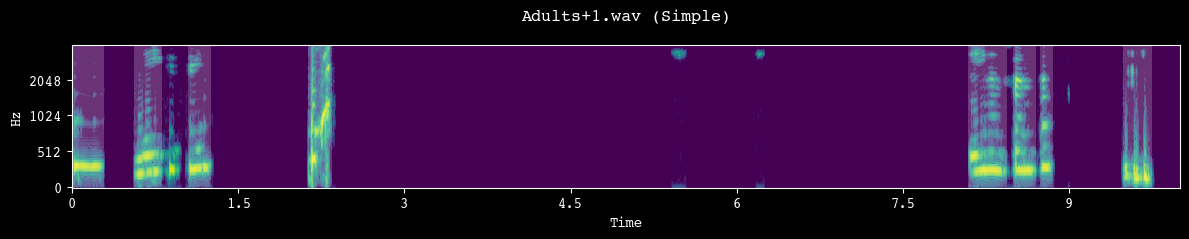
--- Demoing segmentation for: Adults+1.wav ---
Segment: 155.11s - 165.11s
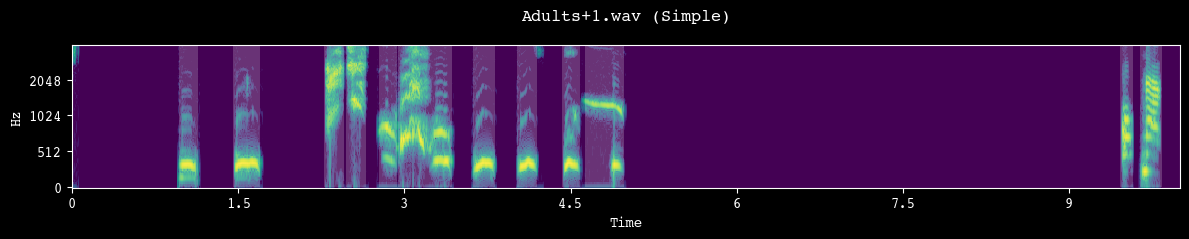
[10]:
# segment units and save spectrograms
unit_df = analyzer.segment_and_create_spectrograms(
processed_dir=processed_dir, h5_path=h5_path, csv_path=csv_path, simple=True
)
--- Found 7 files to segment using simple (amplitude-based) method ---
Segmenting and saving spectrograms: 100%|██████████| 7/7 [00:28<00:00, 4.12s/it]
--- Data preparation complete. Created records for 9744 units ---
Spectrograms saved to /Volumes/Expansion/data/chatter/examples/chimpanzee/spectrograms.h5
Unit metadata saved to /Volumes/Expansion/data/chatter/examples/chimpanzee/spectrograms.csv
Run model#
[11]:
# load segmented units
unit_df = analyzer.load_df(csv_path)
Attempting to load /Volumes/Expansion/data/chatter/examples/chimpanzee/spectrograms.csv...
--- Successfully loaded /Volumes/Expansion/data/chatter/examples/chimpanzee/spectrograms.csv ---
[12]:
# train ae
model.train_ae(unit_df=unit_df, h5_path=h5_path, model_dir=model_dir, subset=0.1)
--- Training on a random subset of 974 units (10.0%) ---
Starting training for 100 epochs using 4 DataLoader workers...
Training model: 100%|██████████| 100/100 [03:38<00:00, 2.19s/it, loss=594.8712]
--- Training complete. Model saved to /Volumes/Expansion/data/chatter/examples/chimpanzee/model/model.pth ---
Loss history saved to /Volumes/Expansion/data/chatter/examples/chimpanzee/model/loss.csv
[14]:
# load trained vae
model = chatter.Trainer.from_trained(config, model_dir)
Instantiating Trainer from pre-trained model at /Volumes/Expansion/data/chatter/examples/chimpanzee/model...
Initializing convolutional variational autoencoder
Using device: mps
Successfully loaded pre-trained model from /Volumes/Expansion/data/chatter/examples/chimpanzee/model/model.pth
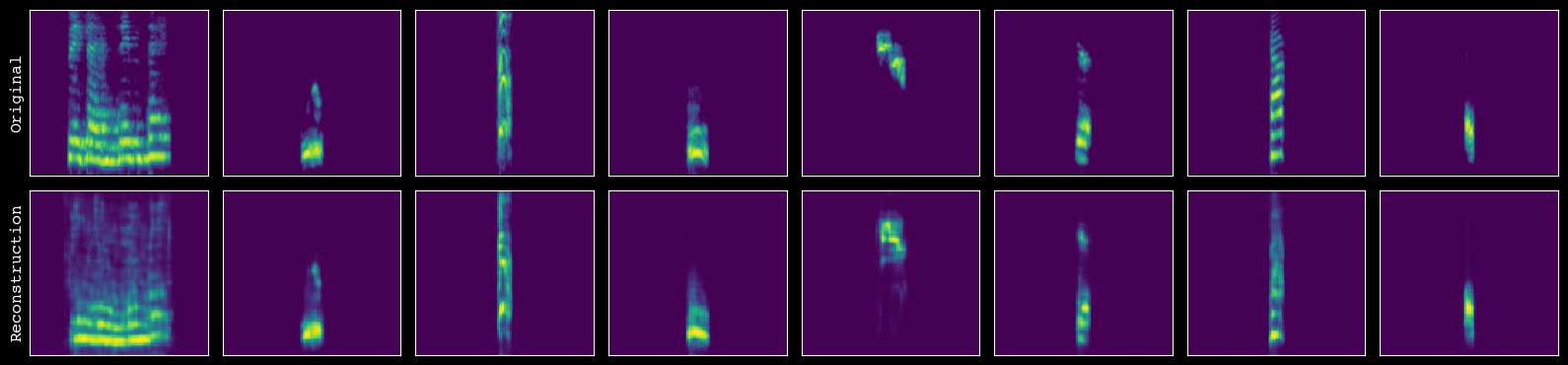
[38]:
# assess the quality of reconstruction
model.plot_reconstructions(unit_df=unit_df, h5_path=h5_path)
[39]:
# export the latent features
output = model.extract_and_save_features(
unit_df=unit_df,
h5_path=h5_path,
model_dir=model_dir,
output_csv_path=output_csv_path,
)
--- Starting feature extraction ---
Successfully loaded pre-trained model from /Volumes/Expansion/data/chatter/examples/chimpanzee/model/model.pth
Extracting features: 100%|██████████| 153/153 [00:06<00:00, 23.61it/s]
--- Pipeline complete. Exported data for 9744 units to /Volumes/Expansion/data/chatter/examples/chimpanzee/output.csv ---
Postprocessing#
[40]:
# load in latent features
output = chatter.FeatureProcessor(analyzer.load_df(output_csv_path), config)
Attempting to load /Volumes/Expansion/data/chatter/examples/chimpanzee/output.csv...
--- Successfully loaded /Volumes/Expansion/data/chatter/examples/chimpanzee/output.csv ---
[41]:
# process output and save
output.run_pacmap()
output.df.to_csv(output_csv_path, index=False)
--- Running PaCMAP dimensionality reduction ---
--- PaCMAP complete ---
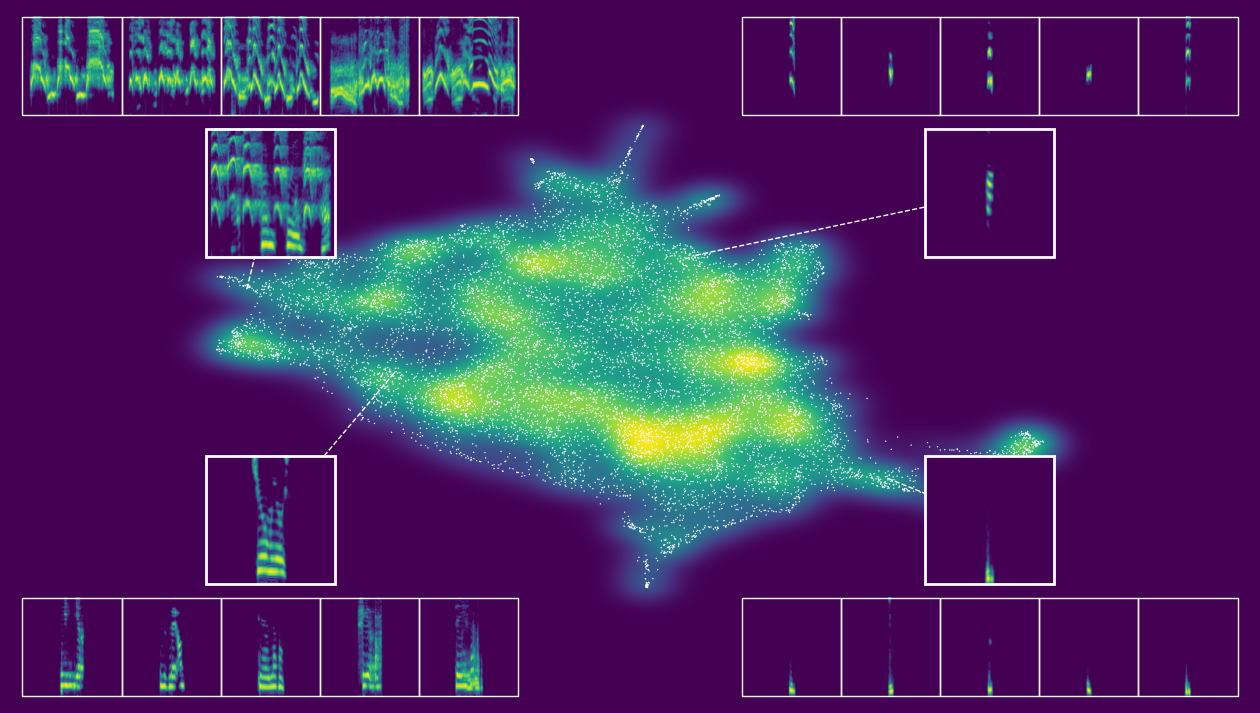
[63]:
# create interactive or static plot
output.static_embedding_plot(
h5_path=h5_path,
seed=111111,
focal_quantile=0.5,
point_size=1,
point_alpha=1,
margin=0.01,
zoom_padding=0.25,
num_neighbors=5,
)
--- Automatically selecting focal points from quadrants with seed 111111 ---
--- Finding nearest neighbors ---
--- Creating the plot ---
--- Plotting density background (using fast 2d histogram) ---
--- Calculating callout positions and adding spectrograms ---
--- Displaying plot ---
[63]:
<chatter.features.FeatureProcessor at 0x341023b60>
